Metabolic perturbation of SARS-CoV-2 as a potential future therapeutic target
Published: 2021-10-07

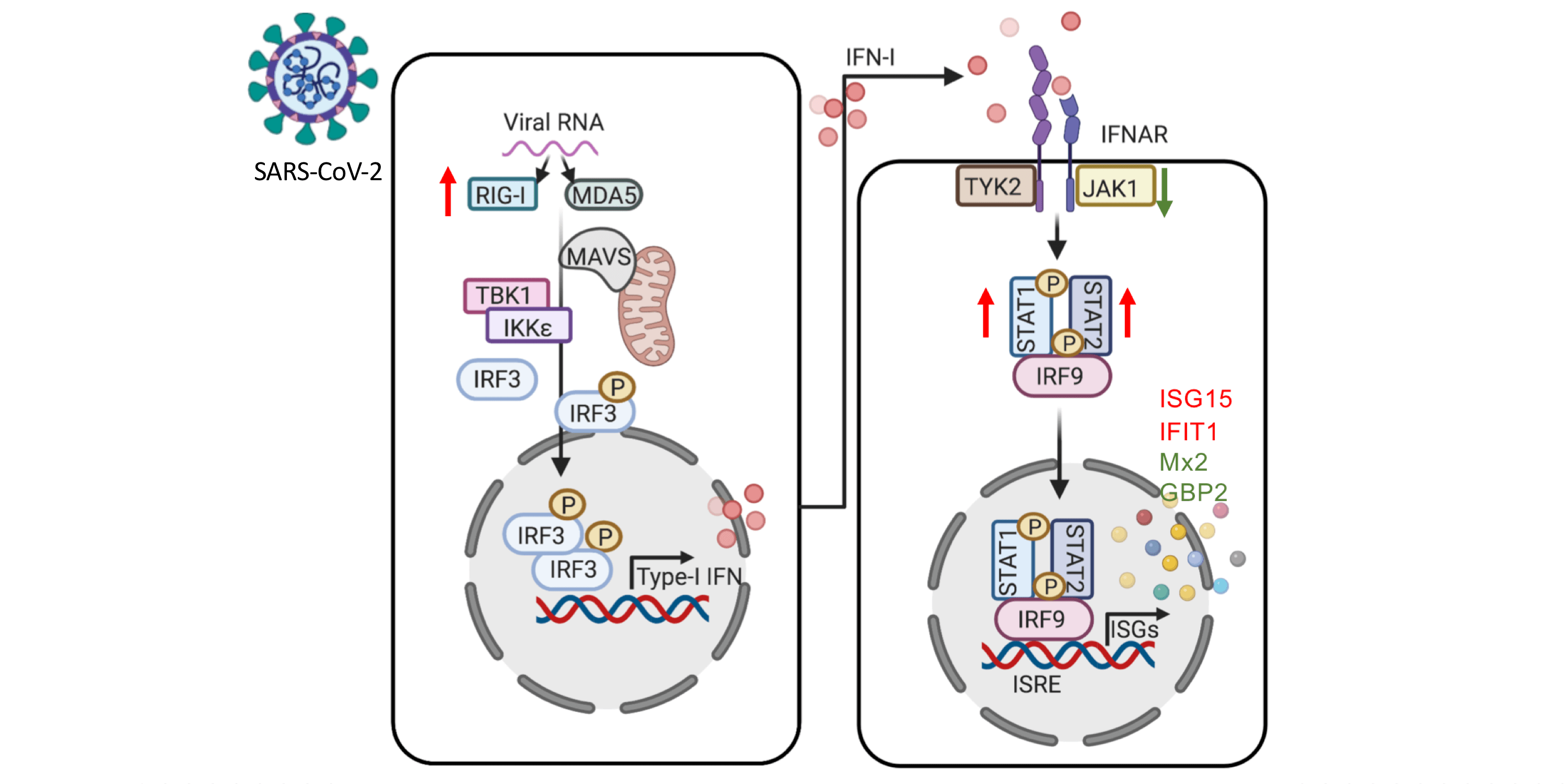
The COVID-19 pandemic has caused public health challenges. The research community has, in record time, increased knowledge and understanding about the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) virus. Recent studies have, for example, found that certain risk factors (e.g. obesity) affect the severity of the resultant COVID-19 disease. Recent observational studies indicated an association between disease severity, and altered energy and lipid metabolism. Metabolic profiling could therefore be one way to identify likely disease severity. One important aspect for future pandemic preparedness is the development of new antivirals. Today´s direct-acting antivirals work by weakening the replication of viruses, future antivirals could instead be based on host-based metabolic strategies to inhibit viruses.
In a recently published (Oct 2021) article in Cellular and Molecular Proteomics, a collaboration between researchers from Karolinska Institutet, Södersjukhuset, National Bioinformatics Infrastructure Sweden (NBIS) at SciLifeLab, Stockholm University, Karolinska University Hospital, Umeå University, and the Indian Institute of Science (First author: Shuba Krishnan, Karolinska Institutet. Corresponding author: Asso. Prof. Ujjwal Neogi, Karolinska Institutet) performed plasma proteomics in order to target 92 plasma proteins that are related to inflammation. The researchers also did plasma untargeted metabolomics followed by immune phenotyping of the lymphocyte and monocyte cell population towards the metabolite transporters.
Krishnan and colleagues used patient-derived multiomics data and in vitro infection assays to get an understanding of the role of key metabolic pathways connected to SARS- CoV-2 reproduction and associated with disease severity. Firstly, the researchers used plasma proteomics to identify distinct clusters of healthy controls (HC, N=31) and COVID-19 PCR positive individuals (hospitalised mild case N=29, hospitalised severe case N=12). The hospitalised patient groups were matched by gender, BMI, and age, the HCs were younger, and had lower BMI. The HC group also contained 10 individuals that were HC-CoV-2 Ab+. The researchers then performed targeted proteomics analyses (secretome); investigating 92 plasma proteins involved in inflammatory responses. The results confirmed those of previous research studies, and also showed that a number of cytokines and chemokines were significantly elevated in COVID-19 patients (mild and severe) compared to HCs (HC and HC-CoV-2 Ab+).
The researchers also performed further studies on the differences between HCs and COVID-19 patients. The results showed that most of the altered proteins were involved in cytokine-cytokine receptor interactions and chemokine signaling, the intestinal network for IgA production, the IL-17 signaling pathway, and the Toll-like receptor signaling pathway. When comparing mild and severe COVID-19 patients, eleven proteins were found to be different, among them were hepatocyte growth factor (HGF), pleiotrophin (PTN), several chemokines (CXCL12, CXCL13, and CCL23), monocyte-chemotactic protein (MCP-3, also known as CCL7), interleukin 12 (IL-12), tumor necrosis factor-like weak inducer of apoptosis (TWEAK), vascular endothelial growth factor (VEGFA), angiopoietin 2 (ANGPT2), and Fas ligand (FASLG). Notably IL-12 was found to be increased in the mild COVID-19 patients group compared to both the severe COVID-19 group and HCs. The results also showed that IL-12 levels were highest in COVID-19 patients compared to HC, though patients with a mild case exihibited higher levels than those with a severe case.
The results from the plasma metabolomic profile were found to follow a similar pattern to the plasma proteomics. The HC and HC-CoV-2 Ab+ groups were combined and compared to the COVID-19 patient group. Metabolites of the central carbon metabolism, including glycolysis, were shown to have distinct changes when comparing both HCs and COVID-19 patients and mild and severe COVID-19 patients.
The researchers also modulated key metabolic pathways, identified using multiomics data, to regulate viral reproduction in vitro. In order to do this, COVID-19 disease severity was characterised using increased plasma levels of glucose and mannose. Using immune phenotyping, the researchers were able to identify altered expression patterns of carbohydrate transporter GLUT1 expression in CD8+ T-cells, intermediate and non-classical monocytes, amino acid transporters, and xCT in classical, intermediate, and non-classical monocytes.
In summary, viruses are known to be able to utilise host metabolic pathways to their replicative advantage. This recent study by Krishnan et al. indicates that the SARS-CoV-2 virus can utilise and rewire pathways that are known to govern central carbon metabolism, which can lead to metabolic toxicity. The researchers found that glycolysis and glutaminolysis are essential for virus replication, and that blocking these metabolic pathways caused significant reduction in virus production. These results highlight host metabolic perturbation as an attractive strategy to limit the replication of the SARS CoV-2 virus and disease severity. The development of new antivirals with novel mechanisms is an important part of fighting future pandemics.
‘Host-directed therapy by altering the metabolic pathway that is required by the virus to replicate, can be an attractive strategy to starve the virus to death and can be novel antiviral strategy not only for SARS-CoV-2 but also for other viruses.’ said Ujjwal Neogi, Asso. Professor at the Department of Laboratory Medicine, Karolinska Institutet.
In order to share their findings openly at an early stage, the researchers shared a preprint of their article in bioRxiv on 21st Feb 2021, the article is now published open access online in Molecular and Cellular Proteomics, available here (accepted 28th Sept 2021). The researchers have also publicly shared the data and code produced in their study.
The research was funded by grants from Swedish Research Council Grants, Karolinska Institutet Stiftelser och Fonder, Åke Wiberg Stiftelse grant, and Ragnar Söderberg Foundation.
Data
- Scaled normalised metabolomics data: available in FigShare.
- Proteomics data: available from the ProteomeXchange Consortium via the PRIDE partner repository (identifier: PXD023760).
- All code used in the study: available in GitHub.
Article
DOI: 10.1016/j.mcpro.2021.100159
Krishnan, S., Nordqvist, H., Ambikan, A. T., Gupta, S., Sperk, M., Svensson-Akusjärvi, S., Mikaeloff, F., Benfeitas, R., Saccon, E., Ponnan, S. M., Rodriguez, J. E., Nikouyan, N., Odeh, A., Ahlén, G., Asghar, M., Sällberg, M., Vesterbacka, J., Nowak, P., Végvári, Á., Sönnerborg, A., Treutiger, C. J., Neogi, U. Metabolic perturbation associated with COVID-19 disease severity and SARS-CoV-2 replication. Molecular & Cellular Proteomics (2021)