Quantitative proteomic provides important knowledge about cellular response against SARS CoV-2
Published: 2021-05-19

The COVID-19 pandemic has caused an unprecedented global public health challenge, as well as challenged societies and economies over the last years. The COVID-19 disease is caused by the SARS-CoV-2 virus, found to be a highly pathogenic coronavirus. Although SARS-CoV-2 is primarily known to affect the lung, cardiovascular, gastrointestinal, liver, neurological, hematological, and skin manifestations have also been documented in the disease pathology. Research suggests that all these anatomical sites could be vulnerable to the virus.
Human cell lines are the so-called first line of experimental models for studies into virus pathogenicity, and an important tool for performing antiviral assays studying different viruses. As SARS CoV-2 is still a new virus, research into basic virology and pathogenesis of the virus in different organs is warranted. Understanding the mechanisms of the viral pathogenesis of a SARS-CoV-2 using research with cell lines can in the future contribute to development of effective antivirals against the infection (see, for example, Appelberg and colleagues, 2020).
In a recent study published in iScience, researchers from Karolinska Institute, Public Health Agency Sweden, Umeå University, and University of Nebraska Medical Centre (first authors: Elisa Saccon, Xi Chen, and Flora Mikaeloff; corresponding author Soham Gupta) compared growth characteristics and studied changes in protein abundance caused by SARS-CoV-2 virus in susceptible cell lines using quantitative proteomics.
Saccon, Chen, Mikaeloff, and colleagues assessed the susceptibility and cytopathogenicity of the first Swedish isolate of SARS-CoV-2 (SWE/01/2020) in cell lines from human lung (Calu-3, A549, 16HBE), colon (Caco2), liver (Huh7), and kidney (293FT) as well as in cell lines from African green monkey kidney cells (Vero-E6) which are often a choice for cell-culture-based infection model for coronavirus research. These cell lines are all commonly used in laboratories for performing antiviral assays.
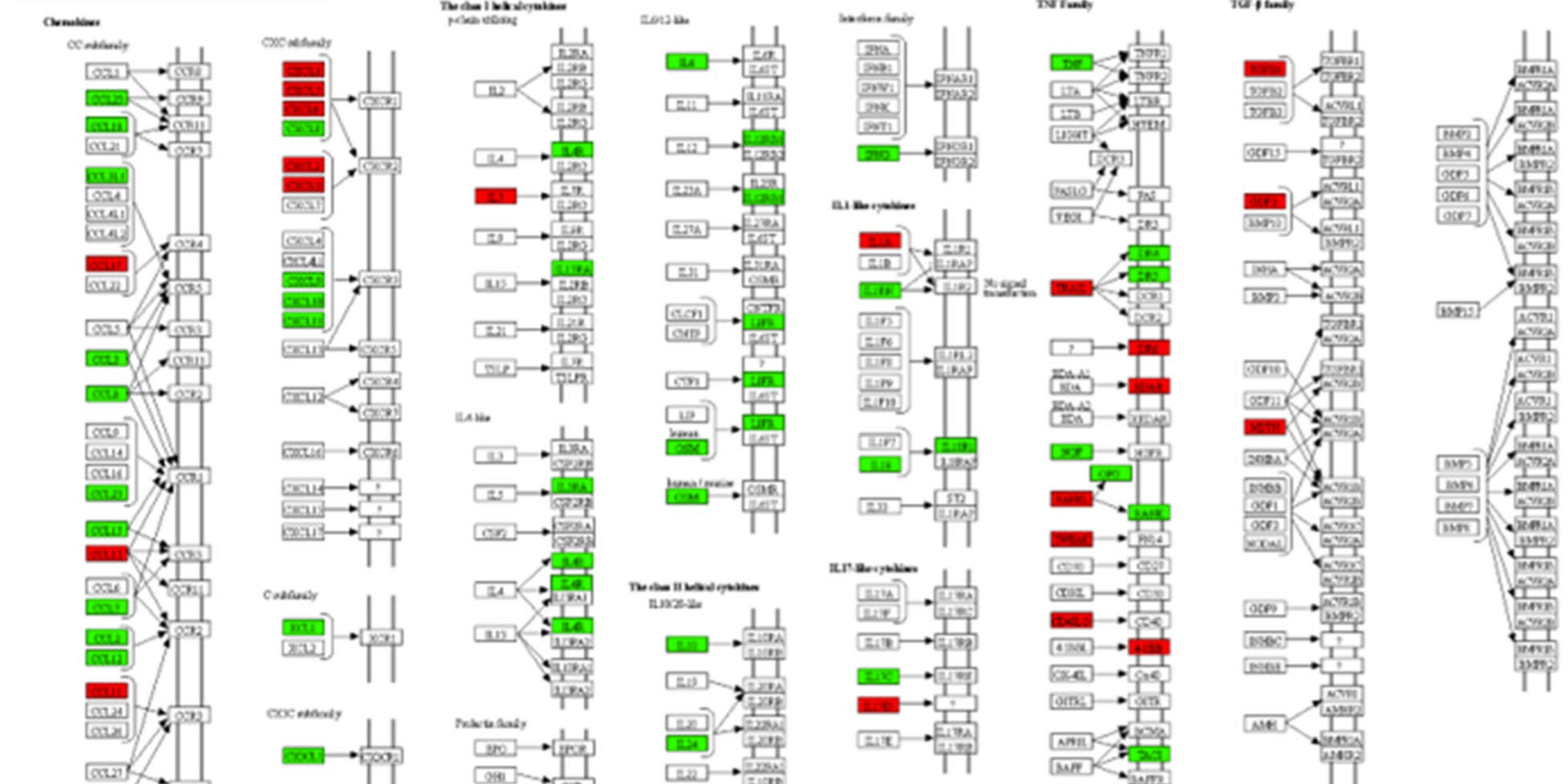
The results provided an overall picture of how the SARS-CoV-2 virus altered the signaling pathways in these cell lines. Calu-3, Caco2, Huh7, and 293FT cell lines were shown to be susceptibility to SARS-CoV-2 (high-to-moderate level). Notably, in Caco2 cells, the SARS CoV-2 virus was found to achieve high titers in the absence of prominent cytopathic effect. In addition, the researchers showed that while there are cell-type specific variations in cellular response, interferon response was found to be commonly dysregulated in Calu-3, Caco-2, and Huh7 cell lines.
The researchers have shared the mass spectrometry proteomics data (raw MS files and search files) openly, depositing them to the ProteomeXchange Consortium via the PRIDE partner repository with the accession number PXD023760. In addition, the researchers have deposited the code openly in GitHub. All the unprocessed and processed proteomics raw data are available from Mendeley Data with DOI 10.17632/tjr7cfhwm7.1.
“In this paper we provide a comprehensive view of how SARS-CoV-2 may behave differently in different human cell-lines by exploring factors that can be related to susceptibility. We believe that our findings will be useful for the researchers to choose a suitable cell model based on the SARS-CoV-2 strain to study pathogenicity, cellular signaling, and antiviral susceptibility. Furthermore, we made all the proteomics data openly available in different repositories that can save a lot of valuable time for fellow COVID-19 researchers,” says the corresponding author of the article, Assistant Professor Soham Gupta.
In summary, Chen, Saccon and Mikaeloff and colleagues identified a number of cell lines of human origin that could prove useful for future studies of the biological properties of SARS-CoV-2. The study also identified type-I interferon regulation during infection in lungs, colon, and liver cell lines which warrant further mechanistic studies in order to identify factors that could be utilized to control the SARS CoV-2 infection. Data produced in this study can provide valuable insight into the cell-type-specific cellular re-organization caused by SARS-CoV-2 and provide important clues to guide future studies.
The researchers within this study were supported by grants from the Swedish Research Council, Karolinska Institutet Foundations and Funds, Åke Wibergs Stiftelse, Innovative Medicines Initiative 2 Joint Undertaking, European Union’s Horizon 2020 research and innovation program, EFPIA, and the Swedish Cancer Society.
Data
- Mass spectrometry proteomics data: ProteomeXchange Consortium, accession number PXD023760
- Code: github.com/neogilab/COVID_cell_lines
- Unprocessed and processed proteomics raw data: Mendeley Data, DOI: 10.17632/tjr7cfhwm7.1
Article
DOI: 10.1016/j.isci.2021.102420
Saccon E., Chen X., Mikaeloff F., Rodriguez J.E., Szekely L., Vinhas B.S., Krishnan S., Byrareddy S.N., Frisan T., Végvári Á., Mirazimi A., Neogi U., Gupta S. Cell type resolved quantitative proteomics map of interferon response against SARS-CoV-2. iScience 24 (5) 102420